Perineural Invasion is a Poor Prognostic Factor for Sinonasal Squamous Cell Carcinoma
Abstract:
Cancer, a multifaceted disease, arises from a complex
interplay of genetic mutations, epigenetic modifications, and proteomic
alterations that drive tumor progression and therapeutic resistance.
Traditional genomic approaches have identified key oncogenes and tumor
suppressors; however, the functional consequences of these alterations at the
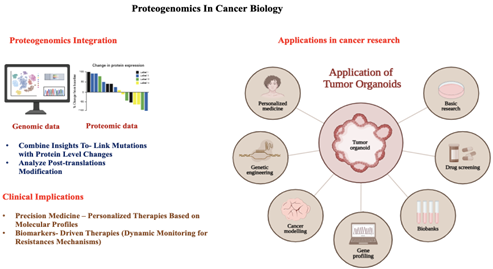
protein level often remain elusive. Proteogenomics, an integrative approach
combining next-generation sequencing and mass spectrometry-based proteomics,
bridges this gap by linking genomic aberrations to proteomic changes, enabling
a deeper understanding of cancer biology. This review highlights the pivotal
role of proteogenomics in unraveling cancer mechanisms, focusing on its
contribution to understanding signaling pathways, post-translational
modifications (PTMs), and tumor heterogeneity. Proteogenomic studies have
elucidated key oncogenic pathways, such as PI3K/AKT/mTOR and MAPK, revealing
how dysregulated proteins and PTMs drive tumor growth and therapeutic resistance.
The approach has also identified novel biomarkers and molecular subtypes across
cancers, facilitating precision medicine. Furthermore, proteogenomics has been
instrumental in addressing therapeutic resistance by uncovering compensatory
mechanisms, clonal evolution, and proteomic adaptations in resistant tumor
cells. Breast cancer and melanoma case studies illustrate its potential in
developing combination therapies to counter resistance. With clinical
applications advancing, proteogenomics holds promise for transforming cancer
treatment through personalized medicine, patient stratification, and
biomarker-driven therapies. Integrating multi-omic data provides a dynamic and
comprehensive view of tumor biology, paving the way for innovative strategies
to improve patient outcomes and combat therapeutic resistance. This review
underscores proteogenomics as a cornerstone in the evolving landscape of
oncology research.
References:
[1]. Balasooriya, E. R., et al., 2024,
Integrating clinical cancer and PTM proteomics data identifies a mechanism of
ACK1 kinase activation. Molecular cancer
research: MCR. 22, 2, 137–151.
[2]. Eroglu, Z., and Ribas, A., 2016.
Combination therapy with BRAF and MEK inhibitors for melanoma: latest evidence
and place in therapy. Therapeutic
advances in medical oncology. 8, 1, 48–56.
[3]. Fujimoto, Y., et al., 2020,
Combination treatment with a PI3K/Akt/mTOR pathway inhibitor overcomes
resistance to anti-HER2 therapy in PIK3CA-mutant HER2-positive breast cancer
cells. Scientific reports. 10, 1,
21762.
[4]. Gerlinger, M., et al., 2012,
Intratumor heterogeneity and branched evolution revealed by multiregion
sequencing. The New England journal of
medicine. 366, 10, 883–892.
[5]. Kannan, B., Pandi, C., Pandi, A.,
Jayaseelan, V. P., & Arumugam, P. 2024, Triggering receptor expressed in myeloid cells 1 (TREM1) as
a potential prognostic biomarker and association with immune infiltration in
oral squamous cell carcinoma. Archives of oral biology, 161,
105926.
[6]. Hanahan, D., and Weinberg, R. A.,
2011, Hallmarks of cancer: the next generation. Cell. 144, 5, 646–674.
[7]. Lei, J. T., and Zhang, B., 2021,
Proteogenomics drives therapeutic hypothesis generation for precision oncology.
British journal of cancer. 125, 1,
1–3.
[8]. Li, Q., et al., 2022, Targeting the
PI3K/AKT/mTOR and RAF/MEK/ERK pathways for cancer therapy. Molecular biomedicine. 3, 1, 47.
[9]. Li, Y., et al., 2023, Proteogenomic
data and resources for pan-cancer analysis. Cancer
cell. 41, 8, 1397–1406.
[10]. Ramarajyam, G., Murugan, R., and
Rajendiran, S. 2024, Network
pharmacology and bioinformatics illuminates punicalagin's pharmacological mechanisms
countering drug resistance in hepatocellular carcinoma. Human Gene, 42,
201328.
[11]. Mertins, P., et al., 2016,
Proteogenomics connects somatic mutations to signalling in breast cancer. Nature. 534, 7605, 55–62.
[12]. Nesvizhskii, A. I., 2014,
Proteogenomics: concepts, applications and computational strategies. Nature methods. 11, 1, 1114–1125.
[13]. Pan, L., et al., 2024, HER2/PI3K/AKT
pathway in HER2-positive breast cancer: A review. Medicine. 103, 24, e38508.
[14]. Rascio, F., et al., 2021, The
pathogenic role of PI3K/AKT pathway in cancer onset and drug resistance: An
updated review. Cancers. 13, 16,
3949.
[15]. Rodriguez, H., et al., 2021, The
next horizon in precision oncology: Proteogenomics to inform cancer diagnosis
and treatment. Cell. 184, 7,
1661–1670.
[16]. Sekaran, S., and Ganapathy, D., 2024,
Tumor-grafted chorioallantoic membrane model: A versatile approach to study
head and neck squamous cell carcinoma physiopathology and therapeutics testing.
Oral Oncology Reports. 10, 100360,
100360.
[17]. Sever, R., and Brugge, J. S., 2015,
Signal transduction in cancer. Cold
Spring Harbor perspectives in medicine. 5, 4, a006098–a006098.
[18]. Tariq, M. U., et al., 2021, Methods
for proteogenomics data analysis, challenges, and scalability bottlenecks: A
survey. IEEE access: practical
innovations, open solutions. 9, (2021), 5497–5516.
[19]. Terekhanova, N. V., et al., 2023,
Epigenetic regulation during cancer transitions across 11 tumour types. Nature. 623, 7986, 432–441.
[20]. Yadalam, P. K., and Ardila, C. M.,
2024, Artificial intelligence-driven multiomics network analysis reveals
resistance mechanisms in oral cancer. International
dental journal. 74, 5, 1180–1181.
[21]. Yip, H. Y. K., and Papa, A., 2021,
Signaling pathways in cancer: Therapeutic targets, combinatorial treatments,
and new developments. Cells (Basel,
Switzerland). 10, 3, 659.
[22]. Zhang, B., et al., 2014,
Proteogenomic characterization of human colon and rectal cancer. Nature. 513, 7518, 382–387.
[23]. Zhang, Y., et al., 2017, A pan-cancer proteogenomic atlas of PI3K/AKT/mTOR pathway alterations. Cancer cell. 31, 6, 820–832.e3.

